Molecular basis of inheritance class 12 NCERT Solution Biology Chapter 6
Molecular basis of inheritance class 12 Intoduction
The molecular basis of inheritance, a pivotal concept in Class 12 biology, delves into the intricate mechanisms governing the transmission of genetic information from one generation to the next. Deoxyribonucleic Acid (DNA), identified as the genetic material, manifests as a double-stranded helix
with specific base pairing dictating its structure. Processes such as DNA replication ensure faithful duplication during cell division, while the genetic code, transcribed into RNA, orchestrates protein synthesis. This fundamental understanding extends to the regulation of gene expression, mutations, and their implications in genetic disorders, providing a foundational framework essential for comprehending the complexities of heredity at the molecular level.
Molecular basis of inheritance class 12 All Scientist Names
- Johann Friedrich Miescher –Isolated DNA From leukocytes called it nuclein”
- Altman–Found it acidic in nature ‘hence he named it Nucleic acid.
- Erwin Chargaff (1951)–Rule of base pairing
- Roslind Franklin and Maurice wilkins (1953)–X-ray diffraction pattern of DNA
- James watson and francis crick (1953)–Molecular structure of DNA
Molecular basis of inheritance class 12 Definition
The molecular basis of inheritance, as elucidated in Class 12 biology, refers to the underlying mechanisms that govern the transfer of genetic information from one generation to the next at the molecular level. Central to this concept is the structure and function of DNA, the hereditary material, which undergoes processes like replication and transcription to ensure faithful transmission and expression of genetic instructions.
The understanding of the molecular basis of inheritance encompasses the decoding of the genetic code, regulation of gene expression, and the impact of mutations on genetic variability and inheritance patterns. This foundational knowledge is essential for comprehending the fundamental principles that underlie the inheritance of traits in living organisms.
Molecular basis of inheritance class 12 Important Notes
- DNA Structure:
- DNA is a double-stranded helix composed of nucleotides.
- Nucleotides consist of a phosphate group, deoxyribose sugar, and nitrogenous bases (adenine, thymine, cytosine, guanine).
- Base pairing occurs through hydrogen bonds (A-T, C-G).
- DNA Replication:
- DNA replication is semi-conservative, with each strand serving as a template.
- DNA polymerase is the enzyme responsible for adding nucleotides during replication.
- Genetic Code and Protein Synthesis:
- Genetic information is transcribed into mRNA in a process called transcription.
- mRNA carries the code from DNA to ribosomes for translation.
- The genetic code is a triplet code, with each triplet (codon) coding for a specific amino acid.
- Types of RNA:
- mRNA carries the genetic code.
- tRNA brings amino acids to the ribosome.
- rRNA is a structural component of ribosomes.
- Regulation of Gene Expression:
- Gene expression is controlled by transcription factors and regulatory proteins.
- It occurs at the transcriptional and post-transcriptional levels.
- Mutations and Genetic Disorders:
- Mutations are changes in the DNA sequence and can be caused by various factors.
- Some mutations may lead to genetic disorders.
- Epigenetics:
- Epigenetic modifications, such as DNA methylation and histone acetylation, influence gene expression without altering the DNA sequence.
- Human Genome Project (HGP):
- The HGP mapped and sequenced the entire human genome, providing valuable insights into the organization and function of genes.
- Applications in Biotechnology:
- Recombinant DNA technology and genetic engineering utilize the principles of the molecular basis of inheritance for various applications.
- Evolutionary Aspects:
- Molecular evidence, such as DNA sequencing, contributes to our understanding of evolutionary relationships among species.
Molecular basis of inheritance class 12 Explanation
In the preceding chapter, you acquired knowledge about inheritance patterns and the genetic foundation underlying those patterns. During Mendel’s era, the nature of the “factors” governing inheritance patterns was unclear.
In the subsequent century, extensive investigations into the putative genetic material led to the realization that DNA, or deoxyribonucleic acid, serves as the genetic material for the majority of organisms. Class XI covered the understanding that nucleic acids consist of nucleotide polymers.
Living systems contain two types of nucleic acids: DNA and RNA. While DNA primarily functions as the genetic material in most organisms, RNA, although acting as genetic material in some viruses, mainly serves as a messenger. RNA also has additional roles, functioning as an adapter, structural molecule, and, in certain cases, a catalytic molecule.
Class XI provided insights into the structures of nucleotides and the linkage of these monomer units to form nucleic acid polymers. This chapter delves into the discussion of DNA structure, its replication, the transcription process for synthesizing RNA from DNA, the genetic code dictating amino acid sequences in proteins, the process of protein synthesis (translation), and the elementary basis of their regulation.
The complete elucidation of the nucleotide sequence of the human genome in the past decade marks a transformative era in genomics. In the following section, we will delve into the fundamentals of human genome sequencing and its consequential implications.
To initiate our exploration, let’s first comprehend the structure of the most captivating molecule in living systems—DNA. Subsequent sections will unravel why DNA stands as the most abundant genetic material and elucidate its relationship with RNA.
5.1 THE DNA
DNA constitutes a lengthy polymer of deoxyribonucleotides, with its length defined by the number of nucleotides or base pairs. This characteristic varies among organisms; for instance, the bacteriophage φ ×174 comprises 5386 nucleotides,
Bacteriophage lambda has 48502 base pairs, Escherichia coli has 4.6 × 10^6 base pairs, and the haploid content of human DNA amounts to 3.3 × 10^9 base pairs. The discussion will now shift to the structure of this extensive polymer
5.1.1 Structure of Polynucleotide Chain
Recapping the chemical structure of a polynucleotide chain (DNA or RNA), a nucleotide comprises three components: a nitrogenous base (Adenine, Guanine, Cytosine, Uracil, or Thymine), a pentose sugar (ribose for RNA and deoxyribose for DNA), and a phosphate group.
Nitrogenous bases form nucleosides through N-glycosidic linkage to the pentose sugar’s 1′ C. The nucleoside, coupled with a phosphate group through a phosphoester linkage to the 5′ C of the pentose sugar, results in the formation of a nucleotide (or deoxynucleotide based on the sugar type).
The connection of two nucleotides through a 3′-5′ phosphodiester linkage forms a dinucleotide, and additional nucleotides join in a similar manner to create a polynucleotide chain. The polymer’s end thus formed possesses a free 3′ hydroxyl group.
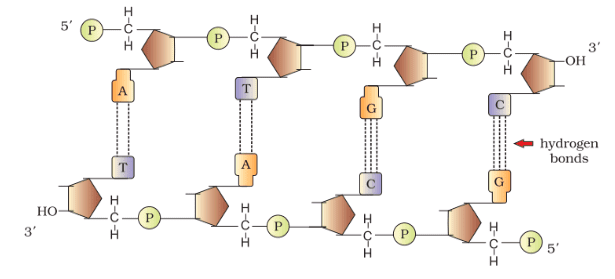
The phosphate moiety at the 5′-end of the sugar defines the 5’-end of the polynucleotide chain, while at the opposite end, the sugar features a free OH group at the 3’C position, referred to as the 3′ -end. The backbone of a polynucleotide chain is constituted by sugars and phosphates,
and the nitrogenous bases linked to the sugar moiety project from the backbone. In RNA, each nucleotide residue possesses an additional –OH group at the 2′ -position in the ribose, and uracil replaces thymine.
DNA, initially identified as an acidic substance in the nucleus by Friedrich Meischer in 1869 and termed ‘Nuclein,’ remained structurally elusive for a significant period due to technical challenges in isolating the intact polymer. In 1953,
James Watson and Francis Crick, building on X-ray diffraction data from Maurice Wilkins and Rosalind Franklin, proposed the famous Double Helix model for DNA’s structure.
This model highlighted the base pairing between the two strands of polynucleotide chains, a concept supported by Erwin Chargaff’s observation that the ratios of Adenine to Thymine and Guanine to Cytosine are constant in double-stranded DNA.
The complementary nature of the polynucleotide chains means that if the sequence of bases in one strand is known, the sequence in the other strand can be predicted.
When each strand from a DNA acts as a template for the synthesis of a new strand, the resulting double-stranded DNA molecules are identical to the parental DNA. The salient features of the Double-helix structure include two polynucleotide chains with an anti-parallel polarity,
base pairing through hydrogen bonds (Adenine with Thymine and Guanine with Cytosine), and a right-handed coiling with a pitch of 3.4 nm and approximately 10 base pairs in each turn.
Consequently, the distance between a base pair (bp) in a helix is approximately 0.34 nm. The stacking of one base pair’s plane over the other in the double helix, along with hydrogen bonds, contributes to the stability of the helical structure (Figure 5.3).
When comparing the structures of purines and pyrimidines, one can discern why the distance between two polynucleotide chains in DNA remains nearly constant.
The proposition of a double helix structure for DNA, with its simplicity in explaining genetic implications, proved to be revolutionary. Shortly thereafter, Francis Crick formulated the Central Dogma in molecular biology, stating that genetic information flows from DNA to RNA to Protein.

In some viruses, the flow of information is in the reverse direction, i.e., from RNA to DNA. The process can be aptly named “Reverse Transcription.”
5.1.2 Packaging of DNA Helix
Considering the distance between two consecutive base pairs as 0.34 nm (0.34×10–9 m), the calculated length of a DNA double helix in a typical mammalian cell (by multiplying the total number of base pairs with the distance between them: 6.6 × 10^9 bp × 0.34 × 10^-9 m/bp) turns out to be approximately 2.2 meters—a length
far exceeding the dimensions of a typical nucleus (approximately 10–6 m). How is such a lengthy polymer packaged within a cell? For instance, if the length of E. coli DNA is 1.36 mm, the number of base pairs in E. coli can be calculated.
In prokaryotes like E. coli, where there is no defined nucleus, DNA is not scattered throughout the cell. Instead, the negatively charged DNA is held by positively charged proteins in a region known as the ‘nucleoid,’ where the DNA is organized into large loops held by proteins.
In eukaryotes, the organization is more intricate. Positively charged, basic proteins called histones play a crucial role. Histones, rich in lysine and arginine, form an octamer known as a histone octamer. The negatively charged DNA wraps around the histone octamer to form a nucleosome, containing 200 base pairs of the DNA helix.
Nucleosomes constitute the repeating unit of a structure in the nucleus called chromatin, visible as thread-like bodies stained in color. When observed under an electron microscope (EM), nucleosomes in chromatin appear as a ‘beads-on-string’ structure. The theoretical estimation of the number of such beads (nucleosomes) present in a mammalian cell is a pertinent consideration.
The chromatin structure, resembling beads-on-string, is further packaged to form chromatin fibers. These fibers are coiled and condensed during the metaphase stage of cell division to ultimately form chromosomes. The higher-level packaging of chromatin necessitates an additional set of proteins collectively referred to as regulatory proteins.
Non-histone Chromosomal (NHC) proteins play a crucial role in chromosomal organization. Within a typical nucleus, certain regions of chromatin are loosely packed, appearing light upon staining, and are referred to as euchromatin.
In contrast, more densely packed chromatin that stains dark is termed heterochromatin. Euchromatin is transcriptionally active, while heterochromatin is inactive.
5.2 THE SEARCH FOR GENETIC MATERIAL
Despite the concurrent discoveries of nuclein by Meischer and the principles of inheritance by Mendel, the recognition that DNA acts as the genetic material required substantial time for discovery and validation. By 1926, the quest to unveil the mechanism of genetic inheritance had progressed to the molecular level.

Earlier findings by Gregor Mendel, Walter Sutton, Thomas Hunt Morgan, and other scientists had narrowed the focus to the chromosomes within the nucleus of most cells. However, the specific molecule serving as the genetic material remained unidentified.
Transforming Principle
In 1928, Frederick Griffith conducted a series of experiments with Streptococcus pneumoniae, the bacterium responsible for pneumonia, leading to a remarkable observation of bacterial transformation.
During his experiments, a living organism (bacteria) underwent a physical change. Streptococcus pneumoniae bacteria, when cultured, yield smooth shiny colonies (S) or rough colonies (R). The S strain has a mucous (polysaccharide) coat, rendering it virulent, while the R strain lacks this coat and is non-virulent.
Mice infected with the S strain succumb to pneumonia, whereas those infected with the R strain do not develop pneumonia. The distinction between these strains marked a pivotal moment in the quest to identify the genetic material.
When Frederick Griffith injected a mixture of heat-killed S (smooth) and live R (rough) bacteria into mice, the mice died, and living S bacteria were recovered from the deceased mice. Griffith concluded that the R strain bacteria had been transformed by some
transforming principle” from the heat-killed S strain, allowing the R strain to synthesize a smooth polysaccharide coat and become virulent. This implied the transfer of genetic material, though the biochemical nature of the genetic material remained undefined from his experiments.
Before the work of Oswald Avery, Colin MacLeod, and Maclyn McCarty (1933-44), the prevailing belief was that the genetic material was a protein. They aimed to determine the biochemical nature of the “transforming principle”
in Griffith’s experiment. Purifying biochemicals (proteins, DNA, RNA, etc.) from the heat-killed S cells, they found that only DNA from S bacteria caused R bacteria to transform. Protein-digesting enzymes (proteases) and RNA-digesting enzymes (RNases) did not affect transformation,
indicating that the transforming substance was not a protein or RNA. However, digestion with DNase inhibited transformation, suggesting that DNA was responsible for the transformation. Despite this evidence, not all biologists were initially convinced.
The conclusive proof that DNA is the genetic material came from the experiments of Alfred Hershey and Martha Chase in 1952. They worked with bacteriophages, viruses that infect bacteria. Bacteriophages attach to bacteria,
and their genetic material enters the bacterial cell. Hershey and Chase aimed to determine whether it was protein or DNA from the viruses that entered the bacteria. Growing some viruses in the presence of radioactive phosphorus and others in the presence of radioactive sulfur,
they found that viruses grown with radioactive phosphorus contained radioactive DNA, while those grown with radioactive sulfur contained radioactive protein. This experiment confirmed that DNA, not protein, is the genetic material.
In the experiments conducted by Alfred Hershey and Martha Chase in 1952, radioactive phages were permitted to attach to E. coli bacteria.
As the infection progressed, the viral coats were removed from the bacteria by agitating them in a blender. Subsequently, the virus particles were separated from the bacteria by spinning them in a centrifuge.
Bacteria infected with viruses containing radioactive DNA were found to be radioactive, indicating that DNA was the material transferred from the virus to the bacteria.
In contrast, bacteria infected with viruses containing radioactive proteins did not exhibit radioactivity. This result strongly suggests that proteins did not enter the bacteria from the viruses. Hence, DNA is conclusively identified as the genetic material passed from the virus to the bacteria (Figure 5.5).
In some viruses, RNA serves as the genetic material, as seen in examples like Tobacco Mosaic viruses and QB bacteriophage. Understanding why DNA is the predominant genetic material while RNA performs dynamic functions such as messenger and adapter roles requires an examination of the chemical differences between the two nucleic acid molecules.
Two notable differences between DNA and RNA include the presence of thymine in DNA instead of uracil and the absence of a 2′-OH group in DNA, making it chemically less reactive and structurally more stable than RNA.
For a molecule to act as genetic material, it must fulfill specific criteria
: (i) It should be able to replicate.
(ii) It should be chemically and structurally stable.
(iii) It should allow for slow changes (mutation) necessary for evolution.
(iv) It should express itself in the form of ‘Mendelian Characters.’
Both DNA and RNA, owing to the rule of base pairing and complementarity, can direct their duplications, meeting the first criterion.
The stability of genetic material is evident in Griffith’s ‘transforming principle,’ where heat did not destroy certain genetic material properties.
DNA’s structural stability is attributed to its complementary strands rejoining under appropriate conditions, contrasting with RNA’s reactivity due to the 2′-OH group at each nucleotide.
Thymine’s presence in DNA further contributes to its stability. Both DNA and RNA can undergo mutations, with RNA being inherently more unstable, mutating at a faster rate. Consequently, RNA-based viruses with shorter life spans mutate and evolve more rapidly.
RNA has the capability to directly code for protein synthesis, allowing for easy expression of characteristics. In contrast, DNA relies on RNA for protein synthesis, as the machinery for protein synthesis has evolved around RNA.
In summary, both RNA and DNA can function as genetic material, but DNA, with its greater stability and dependence on RNA for protein synthesis, emerges as the more suitable genetic material.
Molecular basis of inheritance class 12 Question and Answer
Question:1 If a double-stranded DNA has 20 per cent of cytosine, calculate the per cent of adenine in the DNA.
Answer:
As per Chargaff’s rule, DNA molecules are required to have an equal ratio of purine (adenine and guanine) and pyridine (cytosine and thymine). This is to say that the number of adenine molecules is equivalent to the cytosine molecule.
Percentage of adenosine = percentage of thymine
Percentage of guanine = percentage of cytosine
Hence, according to the law, if the double-stranded DNA has 20% of cytosine, it should have 20% of guanine. Therefore, the percentage of G + percentage of C = 40%
The other 60% indicates both A + T percentage molecules. As adenine and thymine are always found in equal numbers, the adenine content is 30%.
Question:2 List two essential roles of the ribosome during translation.
Answer:
- Decoding mRNA: Ribosomes decode the information carried by mRNA (messenger RNA) molecules. The mRNA serves as a template that carries the genetic code from the DNA in the cell nucleus to the ribosomes in the cytoplasm. The ribosome reads the sequence of codons (three-nucleotide sequences) on the mRNA and interprets it to assemble the corresponding amino acids.
- Peptide Bond Formation: Ribosomes facilitate the formation of peptide bonds between the amino acids. As the ribosome moves along the mRNA, it brings together the appropriate amino acids, positioned on transfer RNA (tRNA) molecules, and catalyzes the formation of peptide bonds between them. This process results in the synthesis of a polypeptide chain, which ultimately folds into a functional protein.
Question:3 Why is the Human Genome project called a mega project?
Answer:
- Scope and Scale: The Human Genome Project aimed to sequence the entire human genome, which consists of approximately 3 billion base pairs of DNA. The sheer size of the genome, combined with the complexity of identifying and mapping all the genes, made it an enormous and unprecedented scientific endeavor.
- Global Collaboration: The Human Genome Project was an international collaboration involving researchers and institutions from around the world. This global cooperation was a distinctive feature of the project, bringing together scientists, institutions, and resources on an unprecedented scale.
- Technological Challenges: At the time the Human Genome Project was initiated in the late 1980s, DNA sequencing technology was in its early stages, and the task of sequencing the entire human genome was technologically challenging. The project required the development and implementation of advanced and high-throughput sequencing technologies.
- Interdisciplinary Nature: The project involved experts from various scientific disciplines, including molecular biology, genetics, bioinformatics, and computational biology. Coordinating efforts across these diverse fields required careful planning and collaboration.
- Significance for Medicine and Science: The completion of the Human Genome Project had profound implications for medicine, biology, and our understanding of human genetics. The project laid the foundation for advancements in personalized medicine, genetic research, and the understanding of various genetic diseases.
- Long Duration: The Human Genome Project spanned over a decade, officially launching in 1990 and reaching its completion in 2003. The sustained effort over this extended period contributed to its characterization as a mega project.
Question:4 What is DNA fingerprinting? Mention its application.
Answer:
Process of DNA Fingerprinting:
- DNA Extraction: The first step involves extracting DNA from a sample, which can be obtained from various sources such as blood, hair, saliva, or other bodily fluids.
- DNA Amplification: Specific regions of the DNA, known as short tandem repeats (STRs) or variable number tandem repeats (VNTRs), are targeted and amplified using polymerase chain reaction (PCR). These regions contain repetitive sequences that vary among individuals.
- Fragment Separation: The amplified DNA fragments are separated based on their size using a technique called gel electrophoresis. This results in a unique pattern of bands representing the individual’s DNA profile.
- Analysis: The DNA profile is then analyzed, and the unique pattern of bands is compared between different individuals. The probability of two individuals having the same DNA profile is extremely low, making it a powerful tool for identification.
Applications of DNA Fingerprinting:
- Forensic Investigations: DNA fingerprinting is widely used in forensic science to identify suspects or link individuals to crime scenes. It has been instrumental in solving criminal cases and exonerating individuals wrongly accused.
- Paternity and Maternity Testing: DNA fingerprinting is employed to establish biological relationships, such as determining parentage in cases of disputed paternity or maternity.
- Immigration Cases: DNA profiling can be used to verify family relationships in immigration cases where individuals need to prove their biological connection to family members.
- Missing Persons and Disaster Victim Identification: In cases of disasters or mass casualties, DNA fingerprinting can aid in the identification of missing persons by comparing DNA profiles of the deceased with samples from relatives.
- Historical and Anthropological Research: DNA fingerprinting is used in genetic studies to trace ancestry, migration patterns, and population genetics. It provides insights into human evolution and historical population movements.
- Wildlife Conservation: DNA fingerprinting is applied in wildlife conservation to study and protect endangered species. It helps in monitoring populations, identifying individuals, and combating illegal wildlife trade.
Molecular basis of inheritance class 12 MCQ Question and Answer
Question:1What is the primary function of DNA in living organisms?
- A) Energy storage
- B) Genetic information storage
- C) Protein synthesis
- D) Cell structure
Question:2 Which enzyme is responsible for unwinding the DNA double helix during replication?
- A) Helicase
- B) DNA polymerase
- C) RNA polymerase
- D) Ligase
Question:3 The building blocks of DNA are known as:
- A) Amino acids
- B) Nucleotides
- C) Polypeptides
- D) Sugars
Question:4 The process of synthesizing mRNA from a DNA template is called
- A) Translation
- B) Transcription
- C) Replication
- D) Mutation
Question:5 What is the role of tRNA in protein synthesis?
- A) Carries amino acids to the ribosome
- B) Transcribes DNA to RNA
- C) Forms the ribosomal subunits
- D) Unwinds the DNA double helix
Question:6 In DNA, adenine pairs with
- A) Cytosine
- B) Thymine
- C) Guanine
- D) Uracil
Question:7 The genetic code is degenerate, meaning
- A) It is unpredictable
- B) It is redundant
- C) It is always changing
- D) It is highly specific
Question:8 Which of the following is responsible for connecting Okazaki fragments during DNA replication?
- A) DNA polymerase
- B) RNA primase
- C) Ligase
- D) Helicase
Question:9 The shape of DNA is often described as
- A) Single-stranded helix
- B) Tetrahedral
- C) Double-stranded helix
- D) Linear
Question:10 Which molecule carries the genetic instructions from the nucleus to the ribosomes for protein synthesis?
- A) mRNA –
- B) tRNA –
- C) rRNA –
- D) DNA
MCQ Answer
| Question No | Answer | Question No | Answer |
| 1 | B | 6 | B |
| 2 | A | 7 | B |
| 3 | B | 8 | C |
| 4 | B | 9 | C |
| 5 | A | 10 | A |
Conclusion
In conclusion, the study of the molecular basis of inheritance in Class 12 biology delves into the intricate mechanisms governing genetic information. It encompasses the structure and function of DNA, the process of replication, transcription, and translation, elucidating the code that guides the synthesis of proteins. Understanding the complementary roles of DNA and RNA,